We are delighted to share our recent article (Allouba et al.) on the influence of ethnicity and consanguinity on the genetic architecture of hypertrophic cardiomyopathy.
Accurate interpretation of genetic variants identified in HCM patients represents a major challenge for diagnosis and implementing precision medicine, especially in understudied populations. Therefore, our aim was to define the genetic architecture of HCM in the largest Middle East and North African (MENA) cohort analysed to date by leveraging an ancestry-matched Egyptian case-control cohort recruited to the Aswan Heart Centre. We also compared genetic data between Egyptian and predominantly European patients to identify patterns of genetic variation that are unique to consanguineous populations of MENA ancestry and are likely to be more important contributors to HCM.
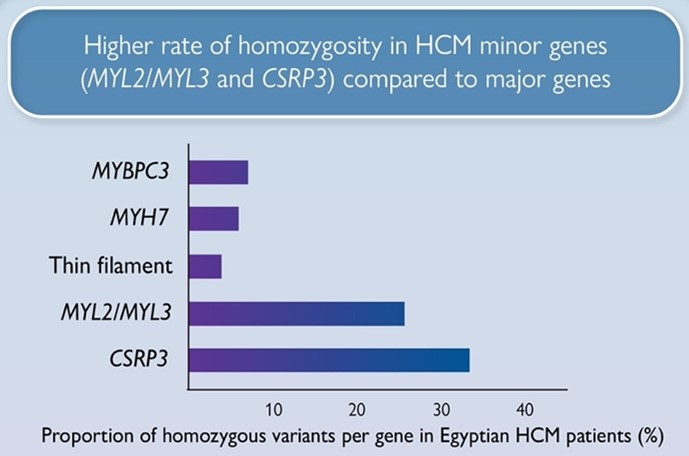
We report a markedly higher rate of homozygosity in HCM minor genes MLC- (MYL2, MYL3) and CSRP3 genes compared to major HCM genes (MYBPC3, MYH7), suggesting these variants have low penetrance in heterozygosity, but contribute to recessive disease. Along with the recently reported recessive HCM gene, TRIM63, these genes could be more relevant to consanguineous populations.
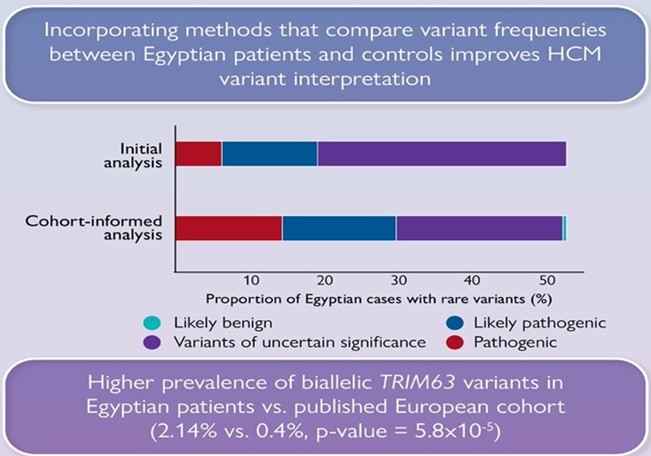
We also show that significantly fewer rare variants detected in Egyptian HCM patients could be classified as (likely) pathogenic compared to Europeans due to the underrepresentation of MENA populations in current HCM databases. Integrating methods that leverage Egyptian controls increased the yield of clinically actionable variants (pathogenic, likely pathogenic) from 19% (initial analysis) to 29.6% (cohort-informed analysis)
Collectively, these findings will enhance the utility of clinical genetic testing for understudied populations as well as our understanding of the genetic aetiology of HCM. Analysis of such highly consanguineous cohorts opens new research avenues for the discovery of novel recessive genes in future exome or genome sequencing studies