The latest ClinGen curation of hypertrophic cardiomyopathy (HCM) genes is now available in pre-print! You can read the full details here.
There are four new definitive HCM genes: ACTN2, CSRP3, FHOD3, and TNNC1.
We have added these to our CardiacG2P dataset. You can check out the latest version of CardiacG2P directly on our github repo, on our dedicated documentation page and on the official EBI G2P page.
Just like in previous releases, this update includes key information on inheritance patterns, allelic requirements, disease-associated variant consequences, and variant classes for each gene-disease pair.
Which Variant Classes Should You Filter For?
The structured dataset lists variant classes that have been reported to cause disease for each gene-disease pair. Importantly, we expect that other variant classes with similar sequence level consequences could also cause disease. For example, if frameshift variants that trigger nonsense-mediated decay (NMD) are known to cause disease, then NMD-triggering stop gained variants may be disease causing as well.
To streamline variant prioritisation, we recommend filtering for all variant classes that map to the disease-associated variant consequences for a given gene-disease pair. Here’s a useful breakdown of variant consequences and their corresponding variant classes:
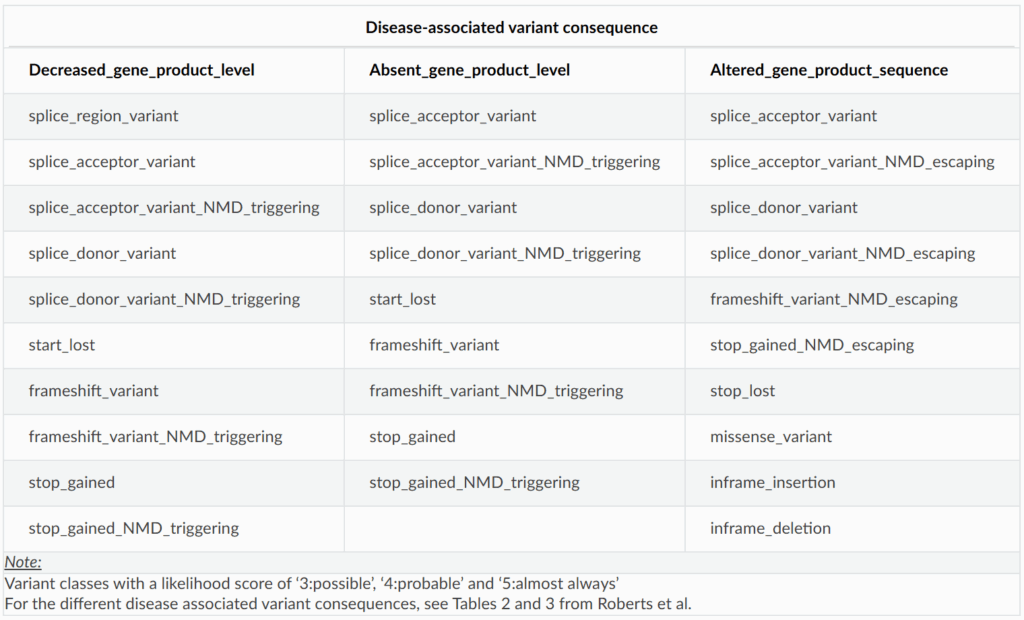
Use this table to guide your variant filtering strategy
For instance, if variants in a gene cause disease through decreased gene product level and stop gained variants have been implicated, you should also filter for variants like splice donor and frameshift variants.
For a more detailed explanation of the disease-associated variant consequences refer to Tables 2 and 3 in Roberts AM et al 2024 (PMID: 37982373). These tables outline the predicted impact of different variant classes at the sequence level.