We are pleased to share our publication (McGurk et al. 2023) in the American Journal of Human Genetics on the penetrance of rare variants in cardiomyopathy-associated genes: a cross-sectional approach to estimate penetrance for secondary findings.
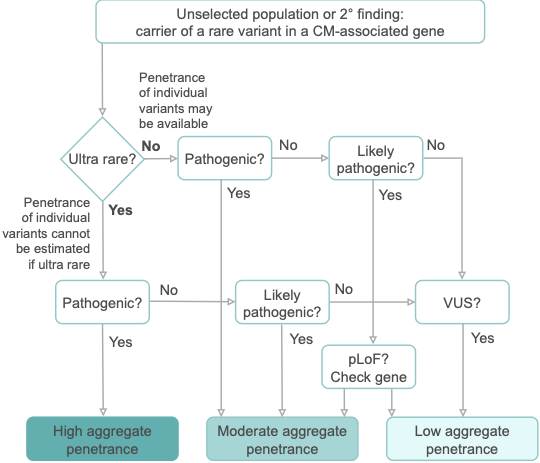
The penetrance of cardiomyopathies (CMs) is incomplete and age-related, and expressivity is highly variable. These features present huge challenges for disease management. In particular, the penetrance of individual variants in CM-associated genes is incompletely characterised and poorly understood, especially when identified in asymptomatic individuals without family history.
With the growing availability of whole exome sequencing in wider clinical settings and consumer-initiated elective genomic testing, the importance of estimating the penetrance of individual variants identified as secondary findings (SFs) to guide intervention is ever-increasing. Genes associated with inherited CMs make up one-fifth of the 78 genes recommended by the American College of Medical Genetics and Genomics (ACMG SF v3.1) for reporting SFs during clinical sequencing. Variant-specific estimates of penetrance are required to appropriately inform clinical practice and to fully utilise genetics as a tool to individualise the risk of developing disease in asymptomatic carriers.
We apply a cross-sectional approach, using a method that compares the allele frequency of individual rare variants in large cohorts of cases and reference populations to estimate penetrance. Sequencing data for 10,400 individuals referred for HCM genetic panel sequencing and 2,564 individuals referred for DCM genetic panel sequencing were included in the analysis. To estimate the prevalence of CMs, a literature review and meta-analysis were undertaken, resulting in prevalence estimates for HCM (1:543; 1:1,300 women, 1:360 men) and DCM (1:220; 1:340 women, 1:160 men).
In aggregate, the penetrance by late adulthood of rare, pathogenic variants (23% for HCM, 35% for DCM) and likely pathogenic variants (7% for HCM, 10% for DCM) was substantial for dominant CM. Penetrance was significantly higher for variant subgroups annotated as loss of function or ultra-rare and for males compared to females for variants in HCM-associated genes.
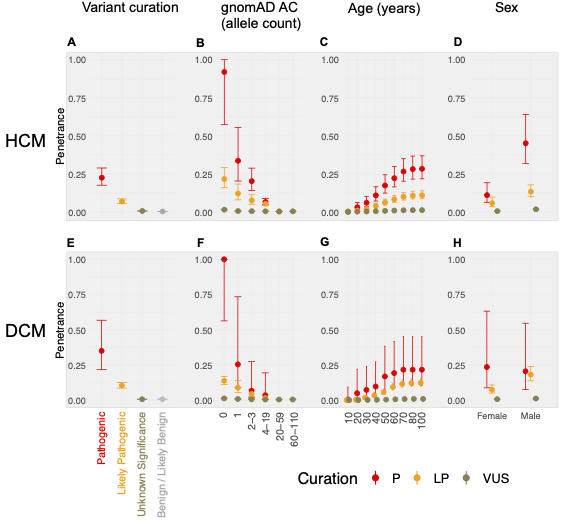
We estimated variant-specific penetrance for 316 recurrent variants most likely to be identified as SFs (51% HCM and 17% DCM cases). 49 variants were observed at least ten times (14% of cases) in HCM-associated genes. Median penetrance was 14.6% (±14.4% SD). We explore estimates of penetrance by age, sex, and ancestry, and simulate the impact of including future cohorts.
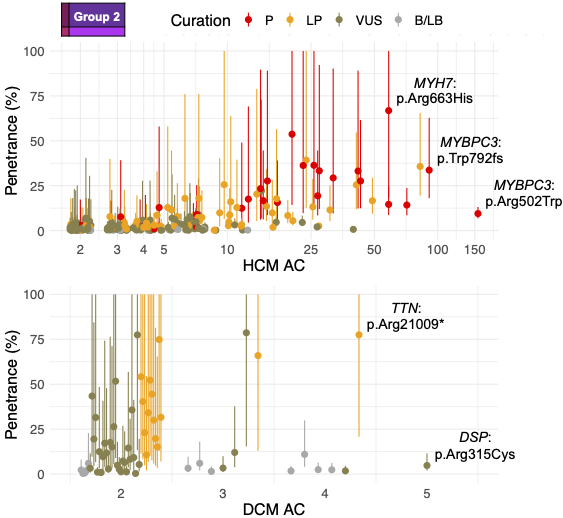
This dataset is the first to report the penetrance of individual variants at scale and will inform the management of individuals undergoing genetic screening for SFs. While most variants had low penetrance and the costs and harms of screening are unclear, some carriers of highly penetrant variants may benefit from SFs.