The Cardiovascular Genetics and Genomics group is delighted to share the systematic review in Wellcome Open Research (McGurk et al. 2022) entitled “Effect of taurine administration on symptoms, severity, or clinical outcome of dilated cardiomyopathy and heart failure in humans: a systematic review”.
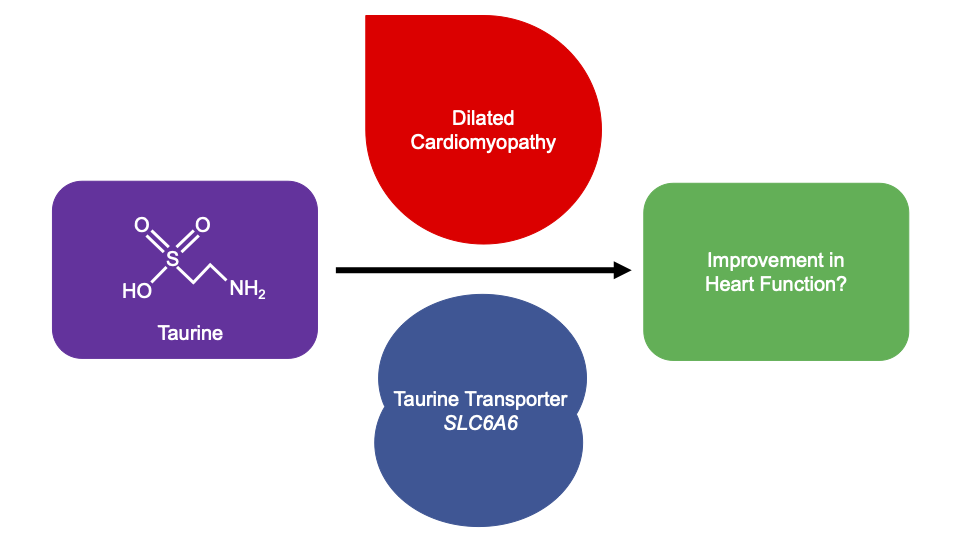
Taurine, 2-aminoethanesulfonic acid, is an essential amino acid found in animal products. Taurine is produced for human consumption as a supplement and ingredient in beverages. Supplementation is a safe, inexpensive, and effective treatment for dilated cardiomyopathy (DCM) in domestic mammals (Pion et al. 1987; Pion et al. 1992; Moise et al. 1991; Keith et al. 2001; Kittleson et al. 1997), however it is currently unlicensed in Europe and the United States for human medical treatment. Recent genome-wide association studies of DCM have identified the locus of the taurine transporter (SLC6A6) (Garnier et al. 2021; Tadros et al. 2021). Additionally, a variant in SLC6A6 has been identified in a consanguineous family with retinal degeneration and mild hypokinetic cardiomyopathy with systolic dysfunction and systolic dilatation of the left ventricle, which corrected with taurine supplementation after 24-months (Ansar et al. 2020). It is unknown whether taurine supplementation can improve human DCM, and furthermore whether improvements are observed when taurine levels are in normal reference ranges.
To assess whether taurine supplementation may be a novel therapeutic option for DCM, we undertook a systematic review. Four electronic databases were searched until 11/03/21. 285 articles were identified, of which eleven met our criteria for inclusion. Taurine supplementation varied across studies; by dose (500 mg to 6g per day), frequency (once to thrice daily), delivery method (tablet, capsule, drink, powder), and duration (2 to 48 weeks). Patient inclusion was all-cause HF patients with ejection fraction (EF) <50% and no study was specific to DCM. While improvements in diastolic and systolic function, exercise capacity, and haemodynamic parameters were described, only EF and stroke volume were measured in enough studies to complete a meta-analysis; the association was not significant with all-cause HF (P<0.05). No significant safety concerns were reported. A formal clinical trial is needed to address whether taurine supplementation is beneficial to the approximately 1/250 individuals with DCM in the population.
How this research came about
The decrease in charity research funding was well publicised in 2020 due to the coronavirus pandemic. I began trying to identify alternative funding sources for future independent research funding. At the time I stayed across the road from a Cats Protection shop. With plenty of time on my hands after work due to UK lockdowns and being new to the field of cardiomyopathy research (without the limits of knowing what is already known), I decided to google whether cats get inherited cardiomyopathies.
Taurine is a regular supplementation for domestic cats presenting with DCM as they lack the ability to create taurine (there are theories around the lack of access of domestic cats to taurine-rich mice). Further research showed that this treatment has been replicated in other domesticated animals. As a geneticist, I did a brief analysis of the taurine metabolic pathway to identify genes where variants may have a potential role in bodily taurine variation, and then assessed the results from a DCM GWAS study we recently published, and I noticed that that one of the hits was in the locus of the taurine transporter (nearest gene LSM3). The lead SNPs are upstream to, and confirmed thyroid eQTLs on GTEx of, the taurine transporter (SLC6A6 or TauT). Without going into detail – the more I dug, the more I found that implicated taurine in DCM and/or altered heart phenotypes.
Why hasn’t a clinical trial for DCM in humans been undertaken?
- taurine tablets are cheap and available (no financial gain?)
- human plasma measurements may be increased by monocyte activation and thus may be inaccurate.
- the mechanism of taurine action is mostly unknown.
- DCM GWAS with large sample sizes have only been published very recently.
- There is a lack of intersection between veterinary medicine and human sciences (diet-associated DCM in dogs has a group on facebook with 114k members…).
Big thank you to the many members of the group for their support – Rachel Buchan and Katherine Josephs provided much needed sanity checks, James Ware supported the idea from the beginning, Melpi Kasapi helped with the systemmatic review, as well as Paul Barton, Stuart Cook, Brian Halliday, Declan O’Regan, Sean Zheng, Ang Roberts, for discussions.
Written by Dr. Kathryn McGurk
https://doi.org/10.12688/wellcomeopenres.17505.1
References
- Ansar M, Ranza E, Shetty M, et al.: Taurine treatment of retinal degeneration and cardiomyopathy in a consanguineous family with SLC6A6 taurine transporter deficiency. Hum Mol Genet. 2020; 29(4): 618–623.
- Garnier S, Harakalova M, Weiss S, et al.: Genome-wide association analysis in dilated cardiomyopathy reveals two new players in systolic heart failure on chromosomes 3p25.1 and 22q11.23. Eur Heart J.; 2021; 42(20): 2000-2011.
- Keith ME, Ball A, Jeejeebhoy KN, et al.: Conditioned nutritional deficiencies in the cardiomyopathic hamster heart. Can J Cardiol. 2001; 17(4): 449–458.
- Kittleson MD, Keene B, Pion PD, et al.: Results of the multicenter spaniel trial (MUST): taurine- and carnitine-responsive dilated cardiomyopathy in American cocker spaniels with decreased plasma taurine concentration. J Vet Intern Med. 1997; 11(4): 204–211.
- McGurk KA, Kasapi M and Ware JS. Effect of taurine administration on symptoms, severity, or clinical outcome of dilated cardiomyopathy and heart failure in humans: a systematic review. Wellcome Open Res 2022, 7:9.
- Moise NS, Pacioretty LM, Kallfelz FA, et al.: Dietary taurine deficiency and dilated cardiomyopathy in the fox. Am Heart J. 1991; 121(2 Pt 1): 541–547.
- Pion PD, Kittleson MD, Rogers QR, et al.: Myocardial failure in cats associated with low plasma taurine: A reversible cardiomyopathy. Science. 1987; 237(4816): 764–768.
- Pion PD, Kittleson MD, Thomas WP, et al.: Response of cats with dilated cardiomyopathy to taurine supplementation. J Am Vet Med Assoc. 1992; 201(2): 275–284.
- Tadros R, Francis C, Xu X, et al.: Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat Genet. 2021; 53(2): 128-134.